Sleeper fish (Bostrychus africanus) are a staple food in West Africa. Harvesting them provides an important source of income for hundreds of communities across the Gulf of Guinea in the Atlantic Ocean. Yet little is known about the genetics of this fish — information that is crucial to safeguarding its genetic diversity, and to enhancing its resilience in the face of climate change and other pressures.
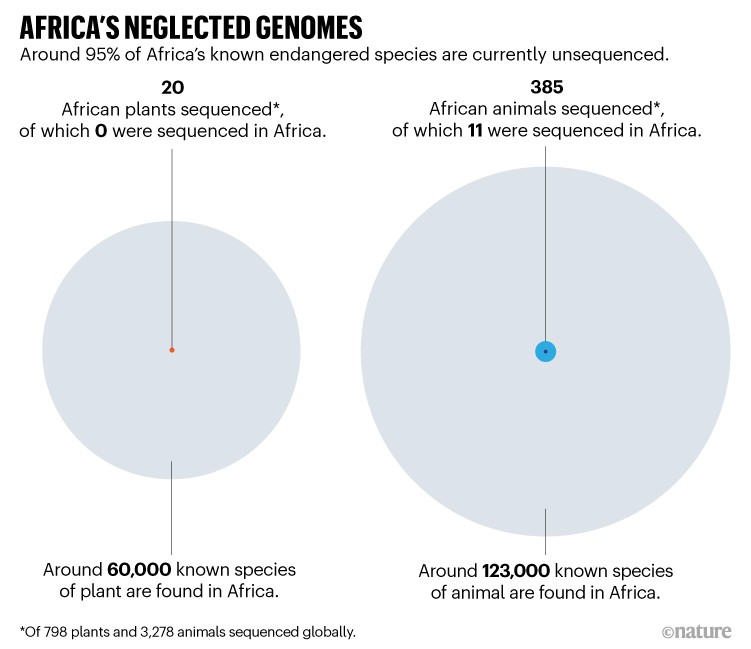
This situation is all too familiar across Africa. Consider orphan crops, which have a crucial role in regional food security, even though they are not typically traded internationally. More than 50% of these have not had their genomes sequenced — from the fluted pumpkin (Telfairia occidentalis) to the marama bean (Tylosema esculentum). The same is true of more than 95% of the continent’s known endangered species (see ‘Africa’s neglected genomes’).
What’s more, by our estimate, around 70% of the 35 or so projects that have focused on studying, conserving or improving biological diversity in Africa over the past 15 years have been led from outside the continent. In fact, among the plant genomes sequenced globally over the past 20 years, almost all of the African species were sequenced elsewhere — mainly in the United States, China and Europe1. This offshoring slows down the much-needed building of expertise and resources in genomics and bioinformatics in Africa (see ‘Africa left out of global genomics efforts’).
The African BioGenome Project (AfricaBP) is an effort to sequence the genomes of 105,000 endemic species: plants, animals, fungi, protists and other eukaryotes. It currently involves 109 African scientists (87 of whom work in Africa) and 22 African organizations.
This store of reference genomes — built in Africa, for Africa — will help plant and animal breeders to produce resilient and sustainable food systems. It will inform biodiversity conservation across the continent. And it will strengthen Africa’s ability to deliver on the goals of the post-2020 global biodiversity framework of the Convention on Biodiversity (CBD). These goals, one of which is to maintain at least 90% of genetic diversity for all known species by 2030, are to be agreed on next month at a meeting in Kunming, China.
An AfricaBP pilot project was launched in June 2021. In this, researchers are sequencing 2,500 indigenous African species, including the Boyle’s beaked blind snake (Rhinotyphlops boylei) from southern Africa and the red mangrove tree (Rhizophora mangle) from Nigeria. They are also mapping out the ethical, legal and social issues raised by a major biodiversity sequencing project — because of cultural sensitivities around certain species, or questions around who has access to the data and who benefits from any resulting discoveries.
For AfricaBP to be scaled up and sustained over the next decade, agencies and organizations need to allocate long-term investments to the project. Such groups include the African Union Commission, national and regional scientific agencies (such as the African Academy of Sciences), and international partners and organizations, including the US National Science Foundation and the UK research funder Wellcome. By our calculations, this will require at least US$100 million per year for the next 10 years (see ‘AfricaBP: structure and costs’).
Some might argue that $1 billion would be better spent on combating malnutrition and disease in impoverished communities across Africa. Yet consider the Human Genome Project, which cost around $3 billion in 2003. By 2019, the human genetics and genomics sector alone was contributing $265 billion annually to the US economy2. Likewise, the World Bank invested millions of dollars in outbreak preparedness from 2017, some of which was used to fund the African Centre of Excellence for Genomics of Infectious Diseases in Ede, Nigeria. This investment meant that Africa was much better equipped to meet the challenges presented by the COVID-19 pandemic.
Species sidelined
Thousands of African species have been ignored by the global genomics community. Only 20 of the 798 plant genomes sequenced globally over the past 20 years are native to Africa1, for example. Yet sub-Saharan Africa alone, which is home to at least 45,000 plant species3, is the second-largest contributor to global plant diversity after South America. Last year, researchers reported that 60% of these species are endemic, and that many could have potential applications in agriculture or drug development4. Evidence suggests, for instance, that African ginger (Siphonochilus aethiopicus) could be used to treat asthma and influenza, among other conditions5,6.
Most of the genomics and bioinformatics expertise that does exist across Africa, including the sequencing facilities, is concentrated in private and non-governmental organizations, such as Inqaba Biotechnical Industries in Pretoria, South Africa, and Redeemer’s University in Nigeria. This means that, although the national research institutes are given the responsibility of setting the country’s scientific agenda, the tools needed to actually improve public health, agriculture and conservation are outside their control7.
AfricaBP will focus on endemic African species that have economic, scientific and cultural significance for African communities.
Sustained government investment in genomics — including the creation of permanent university positions — will help to ensure that African scientists who have received training through African-coordinated genomics projects stay in Africa.
National and regional expansion of tissue-sample collection, taxonomic identification, biobanking of samples and cataloguing of metadata will make it much easier for researchers to monitor species — and ultimately to protect them. Species discovered as a result of the genomics project could be added to the CBD 2030 targets.
Lastly, if the African Union Commission includes AfricaBP in the suite of schemes it is currently backing, the project could enable the commission to achieve at least three of the development goals encapsulated in the African Union Agenda 2063: The Africa We Want. These are: the use of modern techniques and technology to increase agricultural productivity sustainably; the sustainable use of ocean resources to drive economic growth; and the development of environmentally sustainable and climate-resilient economies. (Agenda 2063 is the blueprint for the continent’s transformation into a global powerhouse, as laid out by leaders of the 55 African Union member states in 2013.)
Key priorities
AfricaBP will bring together national and regional institutions, countries and corporations, including already recognized genomics infrastructures, such as the National Institute for Biomedical Research in Kinshasa in the Democratic Republic of the Congo. The project has three main goals.
Improve food systems. The first goal is to provide a resource that enables plant and animal breeders to use various approaches (from conventional breeding to gene editing) to build resilient and sustainable food systems. A 2021 genome analysis8 of 245 Ethiopian indigenous chickens, for instance, revealed the genetic basis of various adaptations that enable the chickens to tolerate harsh environmental conditions (from cold temperatures to water scarcity) — crucial information for poultry producers worldwide. To help achieve this goal, AfricaBP will partner with the African Plant Breeding Academy and the African Animal Breeding Network, both of which were established in the past decade to improve African breeders’ training and research practices.
Improve conservation. The second goal is to make it easier for researchers to identify species and populations that are at risk of extinction, and to design and implement effective conservation strategies. A 2020 study9 on the genetic structure of African savannah elephant populations, for example, revealed that the long-term survival of the elephants requires establishing at least 14 wildlife corridors between 16 of the protected areas in Tanzania. Similarly, a genome study10 of 13 individuals representing 2 subspecies of eastern gorilla showed that inbreeding has led to the purging of severely harmful recessive mutations from one of the subspecies (Gorilla beringei beringei, or mountain gorillas). The accumulation of such damaging mutations in eastern gorillas over the past 100,000 years has reduced their resilience to environmental change and pathogen evolution.
Improve sharing of data and benefits. The third goal is to kick-start a process in which existing multilateral agreements around data sharing are improved and harmonized across the continent — to ensure that the benefits derived from genetic resources are shared equitably across Africa.
In 2010, nations adopted the Nagoya Protocol on Access and Benefits Sharing to ensure that the benefits arising from the use of biological resources are shared fairly. Certainly, any benefit derived from the genetic resources obtained through AfricaBP should be shared by the people of Africa — whether it be a superior strain of drought-resistant sugar beet (Beta macrocarpa Guss) or a new drug derived from the rooibos plant (Aspalathus linearis).
As written, however, the Nagoya Protocol has gaps when it comes to Africa. It fails to take into account the customs and practices of the diverse ethnic groups across the continent. These might not be documented or written into law, but have shaped how people interact with certain plants or animals for hundreds — sometimes thousands — of years. In West Africa, for example, some communities forbid the cutting down or harming of iroko trees, which are thought to have supernatural powers.
There are also inconsistencies in how the Nagoya Protocol is applied in different countries. The African Union guidelines for the implementation of the Nagoya Protocol in Africa states that those countries that are not parties to the Nagoya Protocol should be refused access to the genetic resources of other African member states. But only some countries follow this; South Africa grants non-parties access to the nation’s genetic resources, whereas Ethiopia does not.
Likewise, not all countries require researchers wanting to extract genetic resources to consult community protocols. These include the rules and standards around the handling of biological specimens — as laid out by communities under the guidance of the custodians of customary laws (local chiefs and community heads). These custodians, in turn, work closely with state and national governments; sometimes, community protocols will refer to state, national or international laws. In Benin, for example, such protocols state that researchers cannot enter Gbévozoun forest or take any specimens from it because it houses the deity Gbévo, which protects the community.
Ultimately, it is the responsibility of the African Union Commission to improve and harmonize the treaties and guidelines around data and benefit sharing. Doing this would make it easier for AfricaBP researchers to obtain sampling permits, in accordance with the Nagoya Protocol and material transfer agreements (the legal documents required to send biological materials from one organization to another, or from one country to another).
But AfricaBP will enable the African Union, the CBD and other African agencies, such as the African Academy of Sciences, to integrate genomic information into their policymaking around biological diversity across Africa. This in itself will raise awareness about the Nagoya Protocol, and so encourage greater harmonization in its use.
Furthermore, the 109 scientists championing AfricaBP will coordinate with the African Group of Negotiators on Biodiversity (researchers, policymakers and other stakeholders who represent the continent in CBD negotiations) to ensure that sequencing information is specifically included in the post-2020 global biodiversity framework.
Currently, the Nagoya Protocol specifies that ‘biological samples’ can be exchanged for scientific training or technology transfer. The inclusion of sequencing information would mean that early-career researchers who are members of an Indigenous community, such as the Amhara people in Ethiopia, could negotiate to receive training in genome sequencing and analysis if researchers from South Africa, say, wanted to collect tissue samples from their country.
Lastly, everyone involved in the AfricaBP project — now and over the next decade — will engage local chiefs and other custodians of traditional knowledge in the project from the outset. One way for researchers to engage with local communities or Indigenous peoples is through monthly meetings with government officials involved in Africa’s Access and Benefit Sharing National Focal Points. These individuals are specifically tasked with guiding compliance between the producers of biological resources, such as the Bedouin community in Egypt, and the users of those resources, such as researchers at the Pasteur Institute of Tunis in Tunisia. Another way this could be achieved is through AfricaBP ethics committees surveying thousands of people in a particular community — such as through town-hall meetings, electronic messages or telecommunications.
Making it happen
Since 2009, $22 million has been spent on building bioinformatics capacity across Africa through the Pan African Bioinformatics Network for H3Africa (H3ABioNet) project — including through training 150 researchers in core bioinformatics approaches and technologies. But around 10–15% of the trainees in this Africa-led project have relocated to North America or Europe, and there is no guarantee that they will return. What’s more, H3ABioNet funding winds down this year, and there are few permanent positions for trained bioinformatics personnel in African institutions. Because of this, up to 50% of the researchers who have received training through H3ABioNet could leave Africa.
In the case of AfricaBP, around 600 eligible early-career African researchers (those pursuing PhDs or postdocs) will be granted 3-year fellowships over the next 10 years. They will be able to work with AfricaBP’s global partners11, such as the Wellcome Sanger Institute in Hinxton, UK, through exchange programmes. But they will be based mainly in national and regional AfricaBP facilities, to ensure that any skills they acquire are fed back into the continent.
Cloud-based computing and data storage will need to be coordinated to meet regional needs. Exchange programmes involving AfricaBP partners could help those regions or countries that lack resources; there are currently 87 genomic infrastructures in southern Africa, but only 8 in Central Africa7, for instance. These would be similar to the Newton International Fellowships, which enable early-career researchers from overseas to work for two years at a UK institution.
The 374 state-of-the-art Pacific Biosciences HiFi genome-sequencing machines that currently exist worldwide (as of 31 December 2021) can produce high-quality sequence data for more than 350 species per day12. But although the city of Cambridge, UK, alone has 12 of these machines, there are only 2 in the entire African continent. Building genomics capacity on the ground is a huge challenge in Africa because of the difficulty of transporting intact samples in countries that have poor transport infrastructure and hot climates, and because of Africa’s expensive and low-quality Internet service.
To achieve such a massive sequencing feat, African researchers need state-of-the-art genome technologies. They also need mobile (albeit less accurate13) sequencing technologies that are less reliant on electricity and Internet connectivity, such as the Oxford Nanopore Technologies MinION machine. These are easily transportable and can be used in remote areas; they are roughly the size of a mobile phone13, whereas the Pacific Biosciences HiFi machines are about the size of a household refrigerator.
The 109 scientists spearheading AfricaBP are currently in discussion with leading institutions about the development of mobile sequencing platforms and integrated mobile laboratories. Encouragingly, portable, low-cost computing platforms, such as Raspberry Pi and eBioKit, are already being used in Africa, for instance at Makerere University in Kampala, Uganda, in bioinformatics training programmes.
We ask all African life-science agencies to join AfricaBP. We also ask the African Union Commission and the African Academy of Sciences to provide the core funds — US$100 million per year for the next 10 years. In our view, this investment will be dwarfed by the economic and other pay-offs that will stem from AfricaBP-enabled innovations and discoveries.







More Stories
U.S. charges FTX founder Sam Bankman-Fried with criminal fraud
Time series forecasting with XGBoost and InfluxDB
Full-stack engineering is one-third as good