
Element of a gene is better than none when identifying a species of microbe. But for Rice University personal computer scientists, part was not practically more than enough in their pursuit of a method to recognize all the species in a microbiome.
Emu, their microbial neighborhood profiling program, correctly identifies bacterial species by leveraging long DNA sequences that span the entire size of the gene less than examine.
The Emu project led by computer scientist Todd Treangen and graduate scholar Kristen Curry of Rice’s George R. Brown College of Engineering facilitates the analysis of a vital gene microbiome researchers use to sort out species of micro organism that could be harmful—or helpful—to human beings and the natural environment.
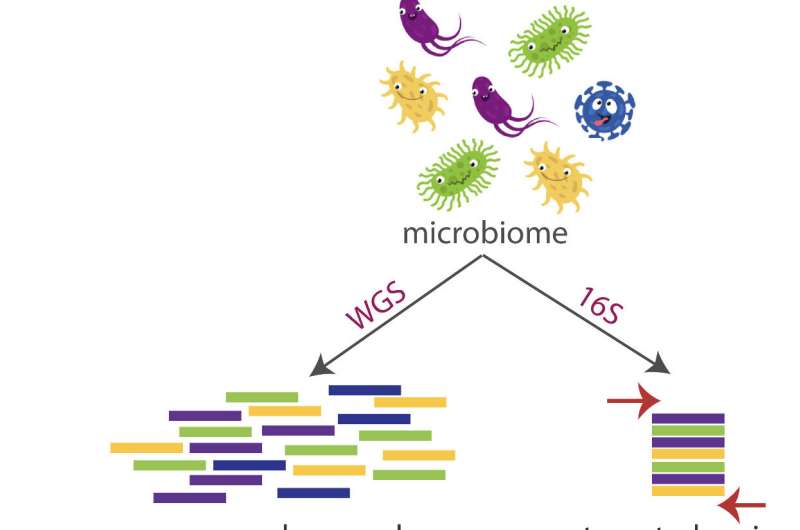
Their goal, 16S, is a subunit of the rRNA (ribosomal ribonucleic acid) gene, whose use was pioneered by Carl Woese in 1977. This region is very conserved in microbes and archaea and also contains variable areas that are significant for separating distinctive genera and species.
“It is normally utilised for microbiome investigation simply because it’s present in all micro organism and most archaea,” reported Curry, in her 3rd year in the Treangen team. “For the reason that of that, there are areas that have been conserved in excess of the a long time that make it quick to target. In DNA sequencing, we will need elements of it to be the exact in all germs so we know what to appear for, and then we want components to be different so we can convey to micro organism apart.”
The Rice team’s study, with collaborators in Germany and at the Houston Methodist Investigate Institute, Baylor Faculty of Medicine and Texas Children’s Clinic, appears in the journal Mother nature Approaches.
“Many years back we tended to target on terrible bacteria—or what we assumed was bad—and we failed to actually care about the others,” Curry reported. “But there’s been a change in the previous 20 yrs to in which we feel maybe some of those other germs hanging out indicate one thing.
“Which is what we refer to as the microbiome, all the microscopic organisms in an natural environment,” she explained. “Typically examined environments incorporate h2o, soil and the intestinal tract, and microbes have proven to have an affect on crops, carbon sequestration and human wellbeing.”
Emu, the title drawn from its task of “expectation-maximization,” analyzes entire-duration 16S sequences from micro organism processed by an Oxford Nanopore MinION handheld sequencer and works by using sophisticated mistake correction to identify species centered on nine unique “hypervariable regions.”
“With prior technology we could only read element of the 16S gene,” Curry described. “It has about 1,500 base pairs, and with quick-examine sequencing you can only sequence up to 25%-30% of this gene. On the other hand, you definitely want the entire-length gene to attain species-amount precision.”
But even the newest technologies just isn’t best, allowing faults to slip into sequences.
“Although error rates have dropped in modern a long time, they can continue to have up to 10% error inside of an personal DNA sequence, when species can be separated by a handful of differences in their 16S gene” claimed Treangen, an assistant professor of personal computer science who specializes in monitoring infectious illness. “Distinguishing sequencing error from accurate distinctions represented the main computational challenge of this investigation challenge.
“A person concern is that a whole lot of the error is nonrandom, this means it can manifest regularly in specific positions, and then get started to search like true discrepancies alternatively of sequencing mistake,” he claimed.
“One more issue is there can be 1000’s of bacterial species in a supplied sample, developing a advanced mixture of microbes that can exist at abundances perfectly beneath the sequencing mistake fee,” Treangen claimed. “This means we are unable to merely rely on advert hoc cutoffs to distinguish signal from error.”
Rather, Emu learns to distinguish between signal and error by comparing a multitude of prolonged sequences, very first towards a template and then from each individual other, refining its mistake-correction iteratively as it profiles microbial communities. In the carried out experiments, phony positives dropped appreciably in Emu in comparison to other approaches when analyzing the exact facts sets.
“Very long-reads signify a disruptive technology for microbiome exploration,” Treangen claimed. “The objective of Emu was to leverage all of the facts contained throughout the whole-duration 16S gene, with out masking nearly anything, to see if we could accomplish far more precise genus- or species-degree calls. And that is particularly what we attained with Emu, many thanks to a fruitful, multidisciplinary collaborative effort and hard work.”
Alexander Dilthey, a professor of genomic microbiology and immunity at Heinrich Heine University, Düsseldorf, Germany, is co-corresponding author of the paper.
Open up-supply software IDs synthetic, obviously transpiring gene sequences
Kristen Curry, Emu: species-degree microbial neighborhood profiling of complete-duration 16S rRNA Oxford Nanopore sequencing facts, Character Methods (2022). DOI: 10.1038/s41592-022-01520-4. www.mother nature.com/content articles/s41592-022-01520-4
Citation:
Emu program utilizes typical gene to profile microbial communities (2022, June 30)
retrieved 6 July 2022
from https://phys.org/news/2022-06-emu-application-popular-gene-profile.html
This document is topic to copyright. Aside from any good working for the purpose of non-public review or investigation, no
section may well be reproduced with no the penned authorization. The articles is supplied for data reasons only.





More Stories
Quantum Space picks up speed with $15M in funding for cislunar space vehicles • TechCrunch
Tender Alerts: What They Are and How They Can Benefit Your Business
The Next Global Economic Revolution